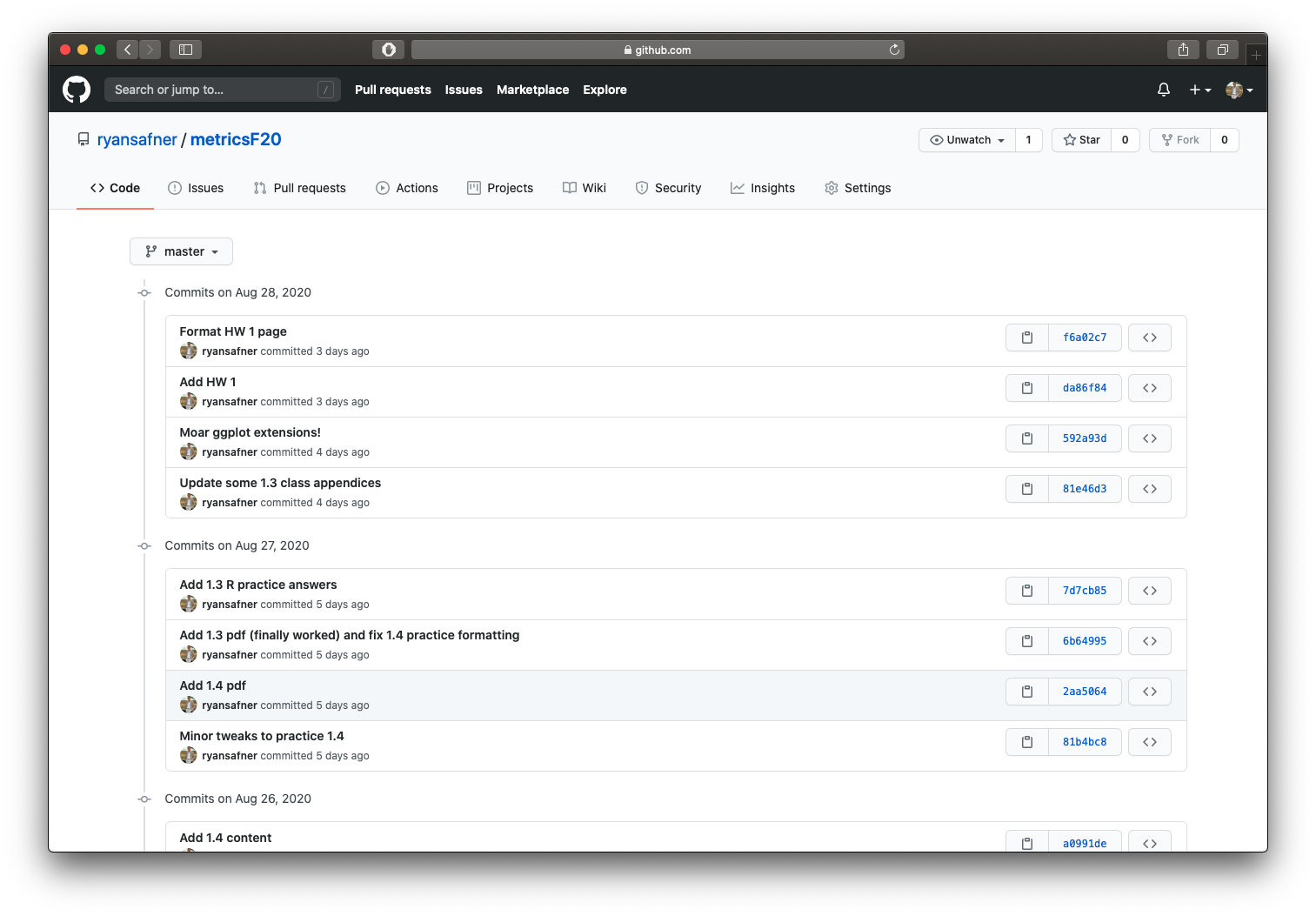
1.5 — Optimize Workflow
ECON 480 • Econometrics • Fall 2021
Ryan Safner
Assistant Professor of Economics
safner@hood.edu
ryansafner/metricsF21
metricsF21.classes.ryansafner.com
Your Workflow Has a Lot of Moving Parts
Writing text/documents
Managing citations and bibliographies
Performing data analysis
Making figures and tables
Saving files for future use
Monitoring changes in documents
Collaborating and sharing with others
Combining into a deliverable (report, paper, presentation, etc.)
The Office Model
The Office Model I
Writing text/documents
Managing citations and bibliographies
Performing data analysis
Making figures and tables
Saving files for future use
Monitoring changes in documents
Collaborating and sharing with others
Combining into a deliverable (report, paper, presentation, etc.)
The Office Model II
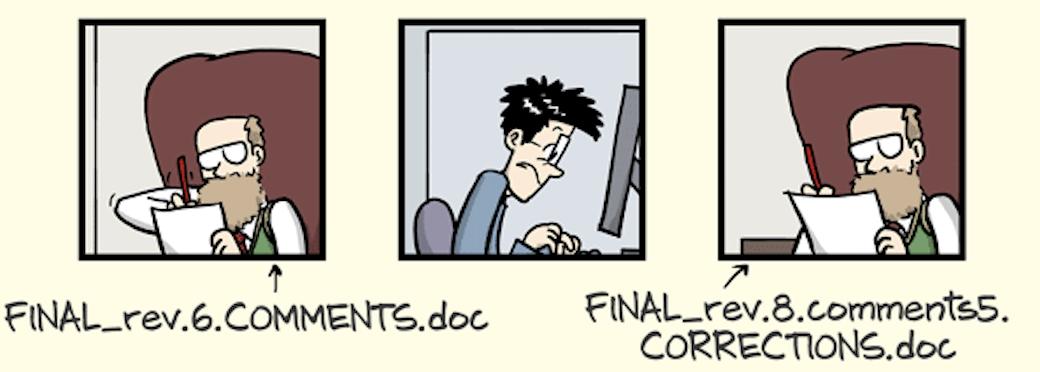
A lot of copy-pasting
A lot of...

The Office Model: A Short Horror Movie
The Office Model: Not Reproducible
...The Rest of the Owl

What I'm About to Show You
This is how I make my...
- Research papers
- Course documents
- Websites
- Slides and presentations
I have not used any MS Office products since 2011 (good riddance!)
This stuff is optional
- If you like your office model, you can keep it
- But this is what most people who take this course continue to use (R is only really if you have data work)
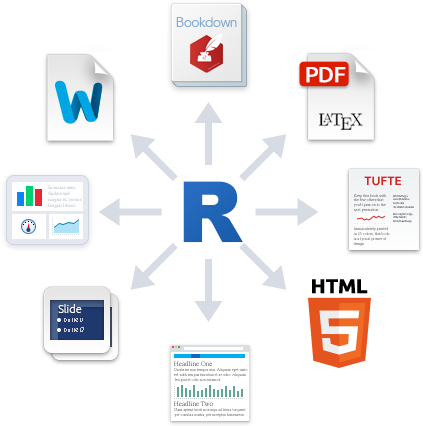
The Plain Text Model
The Plain Text Model I
- Meet
R Markdown, which can do all of this in one pipeline
- Writing text/documents
- Managing citations and bibliographies
- Performing data analysis
- Making figures and tables
- Saving files for future use
- Monitoring changes in documents
- Collaborating and sharing with others
- Combining into a deliverable (report, paper, presentation, etc.)
The Plain Text Model II
Plain text files: readable by both machines and humans
- Understand how a document is structured and formatted via code and markup to text
Focus entirely on the actual writing of the content instead of the formatting and aesthetics
- You can still customize, but with precise commands instead of point, click, drag, guess, pray
The Plain Text Model III
Open Source: free, useable forever, often very small file size
- Proprietary software is a gamble - can you still open a
.docfile from Microsoft Word 1997?
- Proprietary software is a gamble - can you still open a
Automate and Minimize Errors, especially in repetitive processes
Can be used with version control (see below)
Making Your Work Reproducible
One day you will need to quit R, go do something else and return to your analysis the next day. One day you will be working on multiple analyses simultaneously that all use R and you want to keep them separate. One day you will need to bring data from the outside world into R and send numerical results and figures from R back out into the world. To handle these real life situations, you need to make two decisions: What about your analysis is "real", i.e. what will you save as your lasting record of what happened? Where does your analysis "live"?
- Hadley Wickham, R For Data Science
We've talked about
.Rscript files that let you "keep" commandsWhat about output? Must you save and copy/paste to MS Word? No!
Making Your Work Reproducible
R Markdownfile (.Rmd) is the "real" part of your analysis, everything can live in this plain-text file!Document text in
markdownR codeexecuted in "chunks"Plots and tables generated from
R codeCitations and bibliography automated with
.bibfile

R Markdown
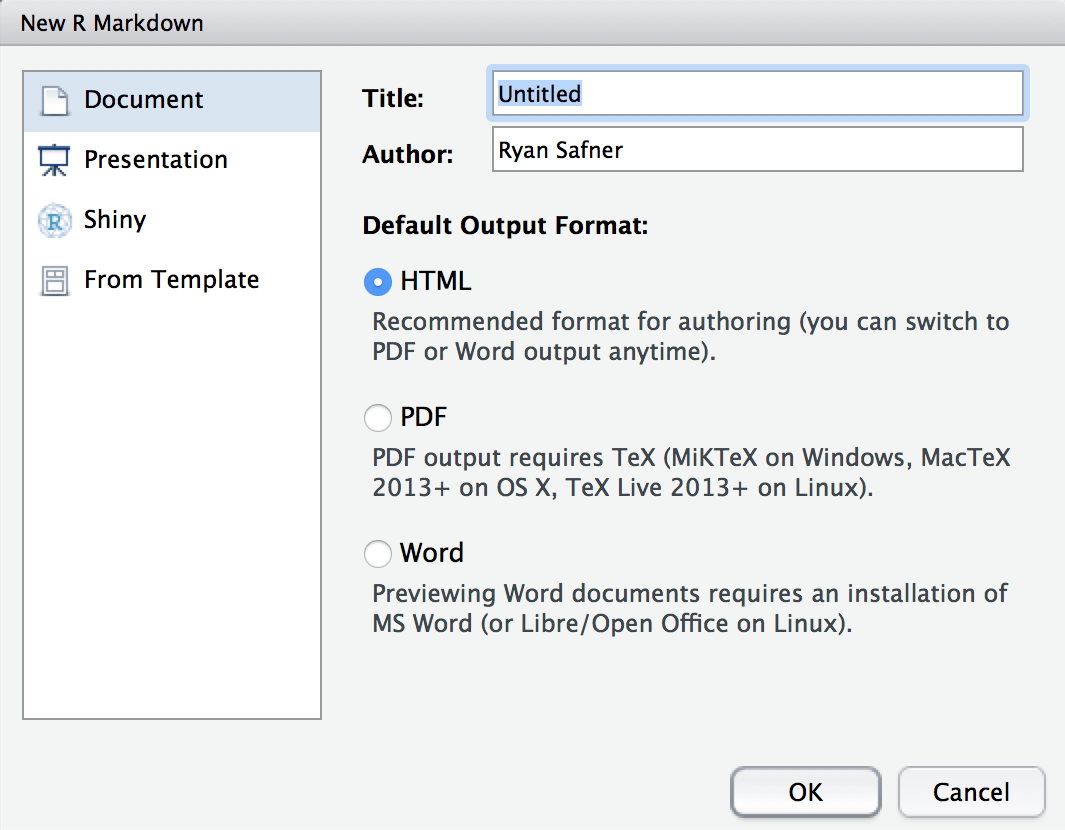
Creating an R Markdown Document I
File -> New File -> R Markdown...
- Outputs:
- Document (what you'll use for most things)
- Presentation (for making slides in various formats)
- Shiny (an html and R based web app, advanced)
- Templates (some built-in, other packages like
rticlesorxaringanadd neat templates)
Creating an R Markdown Document II
File -> New File -> R Markdown...
html: renders a webpage, viewable in any browser- default, easiest to produce and share
- can have interactive elements (gifs, animations, web apps)
- requires internet connection to host and share (you can view offline)
pdf: renders a PDF document- most common document format around
- requires
LaTeXdistribution to render (more on that soon)
word: create a Micosoft Word document- ...if you must

Structure of an R Markdown Document
Entire document is written in a single file1 with three types of content:
YAMLheader for metadataText of the document written with
markdownRchunks for data analysis, plots, figures, tables, statistics, as necessary
1 The one exception is for managing bibliographies, this requires one additional .bib file!
YAML Header I
Top of a document contains the
YAML1 separated by three dashes---above and belowContains the metadata of the document, such as:
title: "My Title"author: "Ryan Safner"date: "`r Sys.Date()`" # here I'm using R code to generate today's date!output: pdf_documentoutputmust be specified, everything else can be left blank, and other options can be added as necessaryIn most cases, you can safely ignore other things in the
yamluntil you are ready
1 YAML stands for "YAML Ain't Markup Language." Nerds love recursive acronyms.
YAML Header: Example from one of my research papers
title: Distributing Patronage^[I would like to thank the Board of Associates of Hood College...]subtitle: Intellectual Property in the Transition from Natural State to Open Access Orderdate: \todayauthor: - Ryan Safner^[Hood College, Department of Economics and Business Administration; safner@hood.edu]abstract: | | "This paper explores the emergence of the modern forms of copyright and patent in ... | *JEL Classification:* O30, O43, N43 | *Keywords:* Copyright, intellectual property, economic history, freedom of the press, economic developmentbibliography: patronage.bibgeometry: margin = 1infontsize: 12ptmainfont: Fira Sans Condensedoutput: pdf_document: latex_engine: xelatex number_sections: true fig_caption: yesheader-includes: - \usepackage{booktabs}R Chunks I
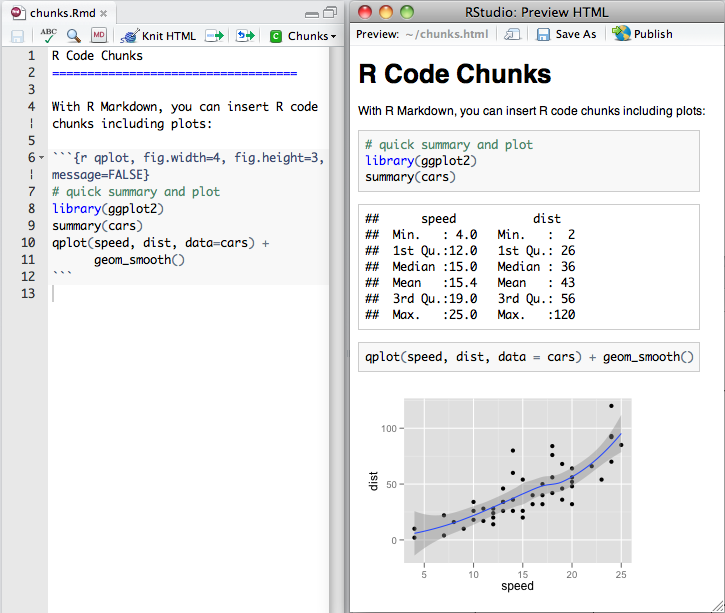
- You can create a "chunk" of
Rcode with three backticks1 above and below your code - After the first pair of backticks, signify the language of the code2 inside braces, e.g:
Input
```{r}2+2 # code goes here!```Output
2+2 # code goes here!## [1] 41 The key to the left of the #1 key on your keyboard.
2 Yes that does mean you can use other coding languages!
R Chunks II
Input
```{r}head(mpg, n=2)```Output
head(mpg, n=2)## # A tibble: 2 × 11## manufacturer model displ year cyl trans drv cty hwy fl class ## <chr> <chr> <dbl> <int> <int> <chr> <chr> <int> <int> <chr> <chr> ## 1 audi a4 1.8 1999 4 auto(l5) f 18 29 p compa…## 2 audi a4 1.8 1999 4 manual(m5) f 21 29 p compa…R Chunks III
Input
```{r}library("ggplot2") # load ggplot2ggplot(data = mpg)+ aes(x = displ)+ geom_histogram()```Output
library("ggplot2") # load ggplot2ggplot(data = mpg)+ aes(x = displ)+ geom_histogram()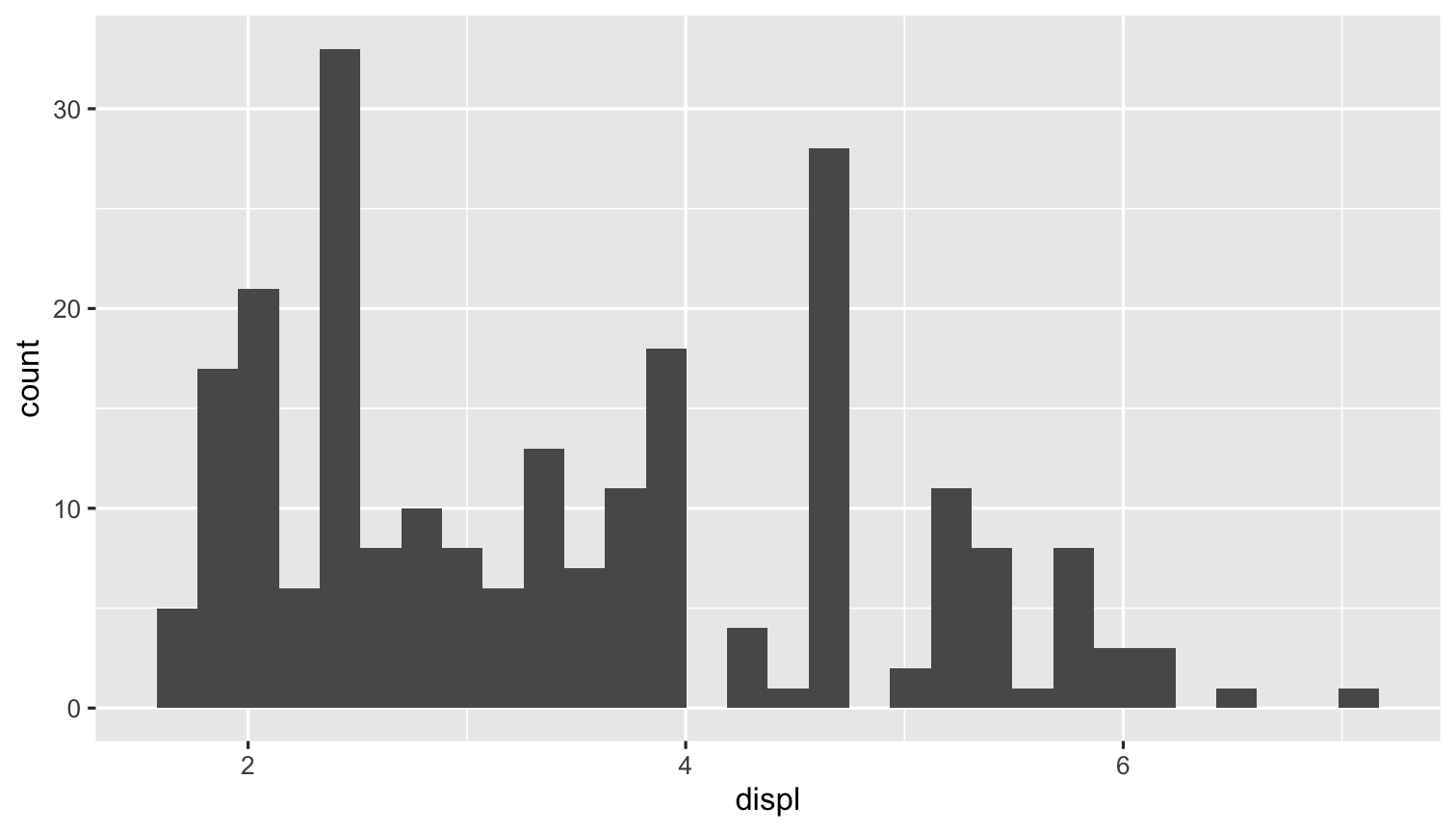
R Chunks Options
You can add additional options inside the
{braces}afterr, some common options:Name: you can name your chunk for further reference later (not required)1
- This is the only option that goes after
rbut before a comma
- This is the only option that goes after
echo- set
=TRUEto display theRcode input - set
=FALSEshows will not show your code
- set
eval- set
=TRUEto run your code =FALSEonly displays your code without running it
- set
fighas a lot of options for displaying plot outputs (fig.height,fig.width,fig.asp, etc)resultswill format the output of a chunk in a certain way (used for advanced things we'll talk about later)
```{r my_cool_chunk, echo=F, warning = F}```R Chunks Options Example
Input
```{r check-data, echo = T}# get top 3 avg displacement by manufmpg %>% group_by(manufacturer) %>% summarize(avg = mean(displ)) %>% arrange(desc(avg)) %>% slice(1:3)``````{r make-plot, echo = F, fig.height=2}ggplot(data = mpg)+ aes(x = displ)+ geom_histogram()```Output
# get top 3 avg engine displacement by manufmpg %>% group_by(manufacturer) %>% summarize(avg = mean(displ)) %>% arrange(desc(avg)) %>% slice(1:3)## # A tibble: 3 × 2## manufacturer avg## <chr> <dbl>## 1 lincoln 5.4 ## 2 chevrolet 5.06## 3 jeep 4.58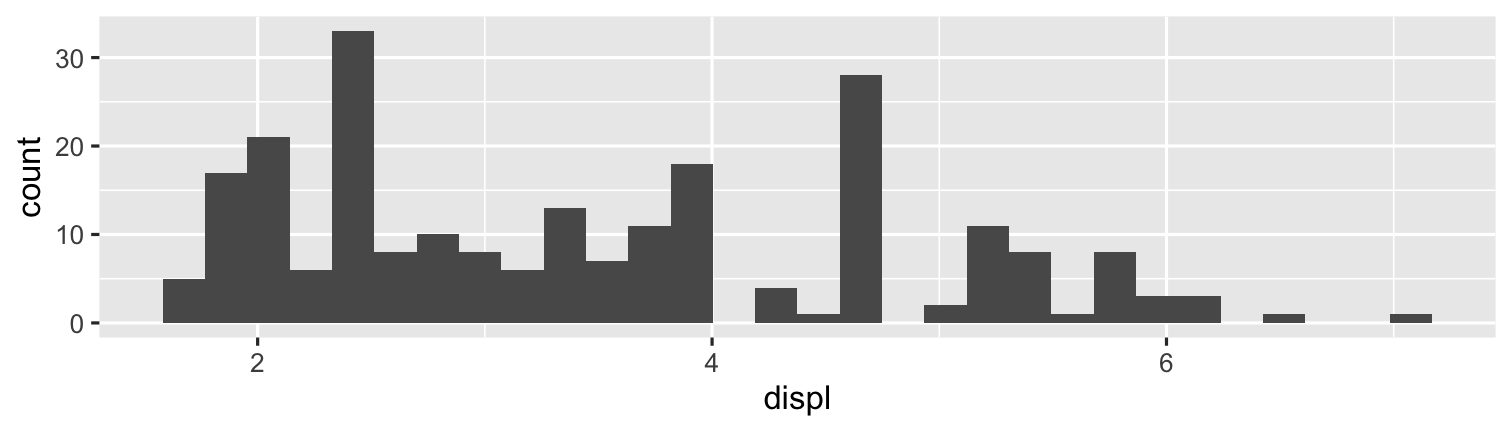
R Chunks Options: Set Defaults
If you want to be fancy, you can set global options that affect all chunks
Use a special named
setupchunk at top (comes in default.Rmdtemplate)- set global options inside the
knitr::opts_chunk$set()command
- set global options inside the
Example on right is what I commonly use in my slides:
- hide all code by default
- hide all messages & warnings
- make figure resolution 3
```{r setup, include=FALSE}knitr::opts_chunk$set(echo = FALSE, message = FALSE, warning = FALSE, fig.retina = 3)```R Inline Code I
If you just want to display some code (or at least format it like code) in the middle of a sentence, place between a single backtick on either side. If I mention
tidyverseorgapminder, it formats the text asin-line code.To actually execute
Rcode to output something in the middle of a sentence, putras the first character inside the backticks, and then run the actual code such as pi is equal to 3.1415927.
Input
pi is equal to `r pi` .
Output
pi is equal to 3.1415927.
R Inline Code II
Input
The average GDP per capita is `r gapminder %>% mean(gdpPercap) %>% round(2)` with a standard deviation of `r round(sd(gapminder$gdpPercap),2)` .
Output
The average GDP per capita is $7215.33 with a standard deviation of $9857.45.
Writing Text with Markdown
- Markdown is a lightweight markup language geared towards HTML (i.e. the internet)
- Markup languages used to add commands about how to display plain text
- Very simple and intuitive
- Write normal text as usual in any word processor
- Change font styling with tags (asterisks):
*italics text*creates italics text**bold text**creates bold text
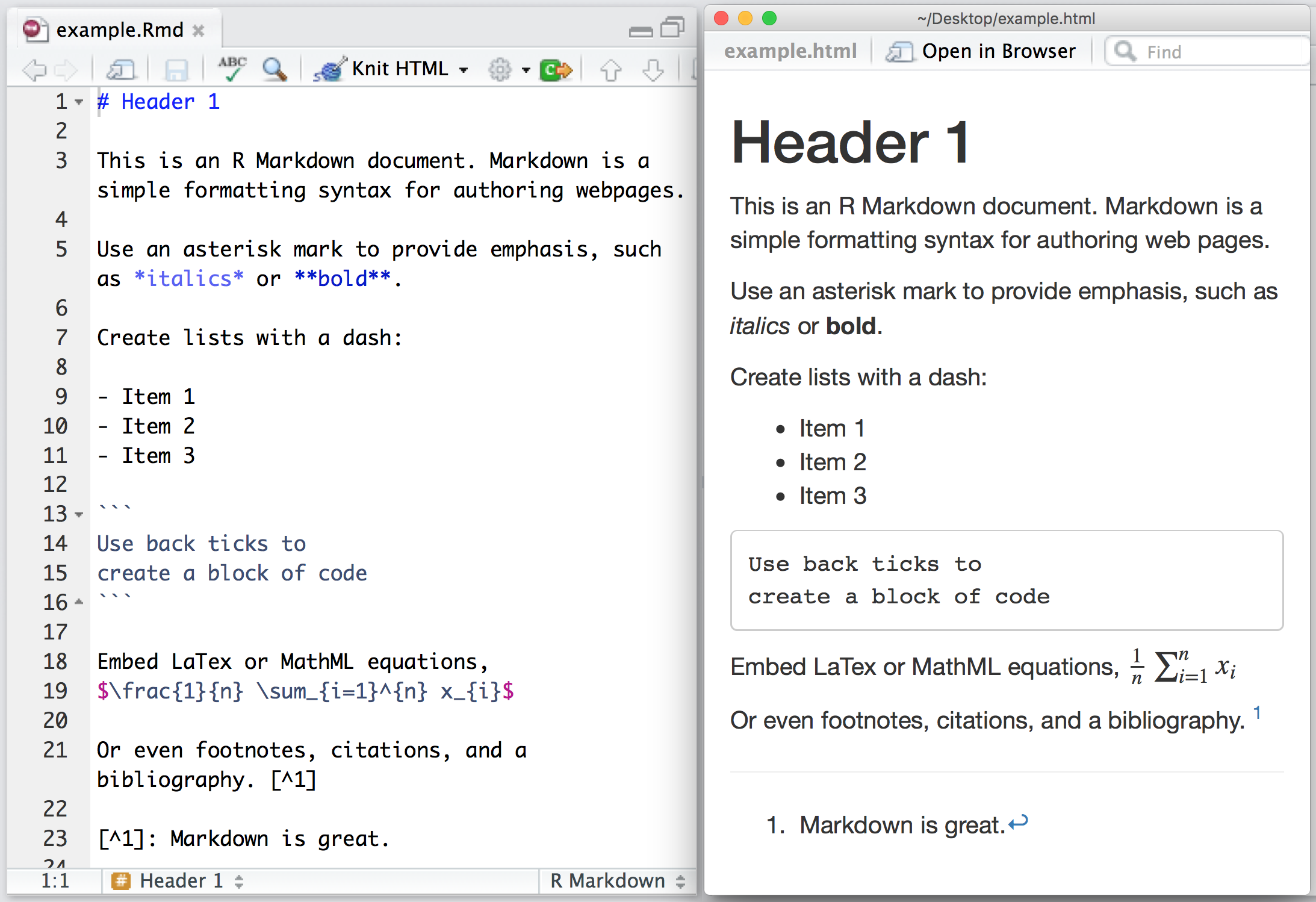
Writing Text with Markdown: Lists
Create an unordered list with lines of (- or + or * ), e.g.:
Markdown is great for taking notes quickly!
Input
- item 1- item 2 - item 2a- item 3Output
- item 1
- item 2
- item 2a
- item 3
Writing Text with Markdown: Headings & Comments
| Markdown | Output |
|---|---|
# Heading 1 |
Heading 1 |
## Heading 2 |
Heading 2 |
### Heading 3 |
Heading 3 |
Comment your code (will not print in output) with <!-- Unprinted comments here --> (this comes from html)
Writing Text with Markdown: Tables
Input
| Header 1 | Header 2 | |----------|----------|| Cell 1 | Cell 2 || Cell 3 | Cell 4 |Output
| Header 1 | Header 2 |
|---|---|
| Cell 1 | Cell 2 |
| Cell 3 | Cell 4 |
- For more complicated tables, there are other packages and techniques
- LaTeX (pdf only)
kableExtrapackagehuxtablepackage (for regression tables)gtpackage
Writing Math I
Add beautifully-formatted math with the
$tag before and after the math, two$$before/after for a centered equationIn-line math example:
$1^2=\frac{\sqrt{16}}{4}$produces 12=√164Centered-equation example:
Input
$$ \hat{\beta_1}=\frac{\displaystyle \sum_{i=1}^n (X_i-\bar{X})(Y_i-\bar{Y})}{\displaystyle \sum_{i=1}^n (X_i-\bar{X})^2} $$
Output
^β1=n∑i=1(Xi−ˉX)(Yi−ˉY)n∑i=1(Xi−ˉX)2
Writing Math II
- Math uses a (much older) language called LaTeX, used by mathematicians, economists, and others to write papers and slides with perfect math and formatting
- I used to use for everything before I found
Randmarkdown - Producing
pdforhtmloutput actually convertsmarkdownfiles into TEX first! (See the process described below) - Much steeper learning curve, a good cheatsheet
- An extensive library of mathematical symbols, notation, formats, and ligatures, e.g.
- I used to use for everything before I found
Writing Math III
| Input | Output |
|---|---|
$\alpha$ |
α |
$\pi$ |
π |
$\frac{1}{2}$ |
12 |
$\hat{x}$ |
ˆx |
$\bar{y}$ |
ˉy |
$x_{1,2}$ |
x1,2 |
x^{a-1}$ |
xa−1 |
$\lim_{x \to \infty}$ |
limx→∞ |
$A=\begin{bmatrix} a_{1,1} & a_{1,2} \\ a_{2,1} & a_{2,2} \\ \end{bmatrix}$ |
A=[a1,1a1,2a2,1a2,2] |
- A great resource: Wikibooks LaTeX Mathematics chapter
Citations, References, and Bibliography
- Manage your citations and bibliography automatically with
.bibfiles - First create a
.bibfile to list all of your references in- You can do this in
Rvia:File -> New File -> Text File(and save with.bibat the end) - See
examplebib.bibin this repository used in this document - At the top of your
YAMLheader in the main document, addbibliography: examplebib.bibsoRknows to pull references from this file - For each reference, add information to a
.bibfile, like so:
- You can do this in
An Example .bib File
@article{safner2016, author = {Ryan Safner}, year = {2016}, journal = {Journal of Institutional Economics}, title = {Institutional Entrepreneurship, Wikipedia, and the Opportunity of the Commons}, volume = {12}, number = {4}, pages = {743-771}}A
.bibfile is a plain text file with entries like thisClasses for
@article,@book,@collectedwork,@unpublished, etc.- Each will have different keys needed (e.g.
editor,publisher,address)
- Each will have different keys needed (e.g.
First input after the
@articleis your citation key (e.g.safner2016)- Whenever you want to cite this article, you'll invoke this key
An Example .bib File
Whenever you want to cite a work in your text, call up the citation key with
@, like so:@safner2016[], which produces (Safner, 2016)You can customize citations, e.g.:
| Write | Produces |
|---|---|
[@Safner2016] |
(Safner, 2016) |
@Safner2016 |
Safner 2016 |
-@Safner2016 |
(2016) |
@Safner2016[p. 743-744] |
(Safner, 2016, p.743-744) |
BibTeX will automatically collect all works cited at the end and produce a bibliography according to a style you can choose
We'll see more when we discuss writing your paper
Reference Management Software
- For more information and examples, see R Studio's R Markdown Guide on Bibliographies
- Lot of programs can help you manage references and export complete
.bibfiles to use withR Markdown
Plain-Text Editors
Markdown files are plain text files and can be edited in any text editor
- something as basic (and boring!) as "Notepad," for example
- many good text editors out there, I like Typora or Ulysses (Mac only) for writing (and previewing) Markdown in a simple interface, with no distractions
Any good editor will have syntax highlighting and coloring when you use tags (like bold, italic,
code, andcode #comments).
R Studio is My Text Editor of Choice
Honestly, I write everything in R Studio's text editor
- Syntax highlighting
- Actually can run R code, autocomplete, etc
- Can render the markdown to an output format: html, pdf, etc.
You can write R code in other text editors, but you can't execute them outside of R Studio (or the command line, but that's too advanced.) Same with actually rendering your markdown to an output (pdf, html, etc)
Tips with Markdown
Empty space is very important in markdown
Lines that begin with a space may not render properly
Math that contains spaces between the dollar-signs may not render properly
Moving from one type of content to another (e.g. a heading to a list to text to an equation to text) requires blank lines between them to work
Compiling Your Documents
knitr
When you are ready, you "compile" your markdown and code into an output format using:
knitr1, an R package that "knits" your R code and markdown.Rmdinto a.mdfile for:pandoc is a "swiss-army knife" utility that can convert between dozens of document types
All you need to do is click the
Knitbutton at the top of the text editor!
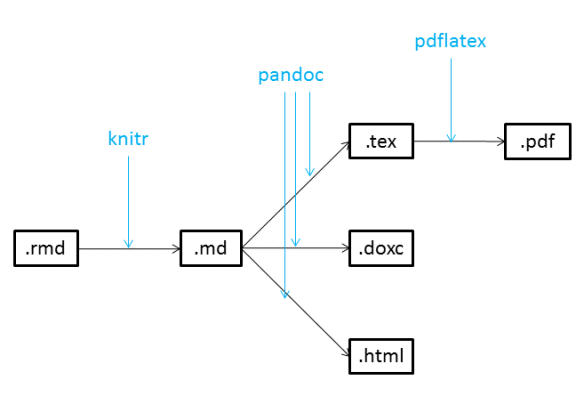
knitr also relies on the rmarkdown package, which will probably be installed when you first knit.R Projects
R Projects I
- A
R Projectis a way of systematically organizing yourRhistory, working directory, and related files in a single package - Can easily be sent to others who can reproduce your work easily
- Connects well with version control software like GitHub
- Can open multiple projects in multiple windows
R Projects II
- Projects solve all of the following problems:
- Organizing your files (data, plots, text, citations, etc)
- Having an accessible working directory (for loading and saving data, plots, etc)
- Saving and reloading your commands history and preferences
- Sending files to collaborators, so they have the same working directory as you
Creating a Project I
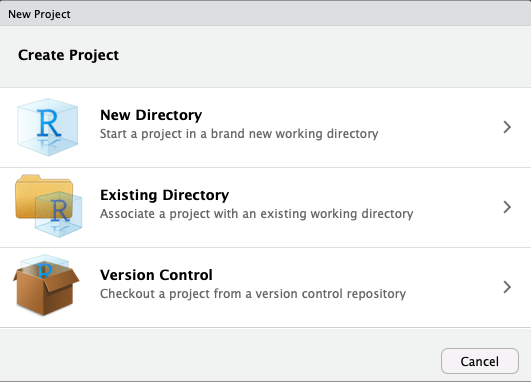
Creating a Project II
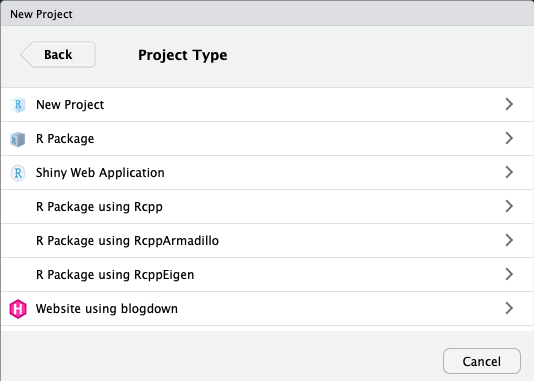
In almost all cases, you simply want a
New ProjectFor more advanced uses, your project can be an
R Packageor aShiny Web ApplicationIf you have other packages that create templates installed (as I do, in the previous image), they will also show up as options
Creating a Project III
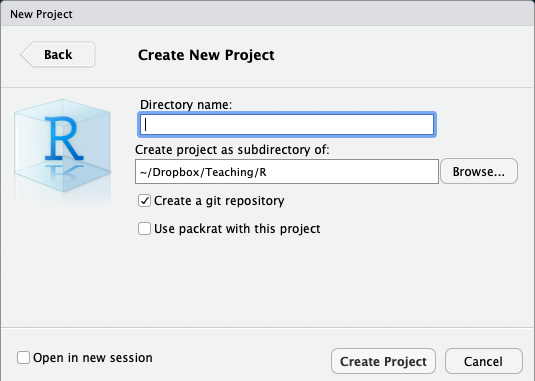
Enter a name for the project in the top field
- Also creates a folder on your computer with the name you enter into the field
Choose the location of the folder on your computer
Depending on if you have other packages or utilities installed (such as
git, see below!), there may be additional options, do not check them unless you know what you are doingBottom left checkbox allows you to open a new instance (window) of
Rjust for this project (and keep existing windows open)
Projects

Switch between each project (Window) on your computer (this is on a Mac).
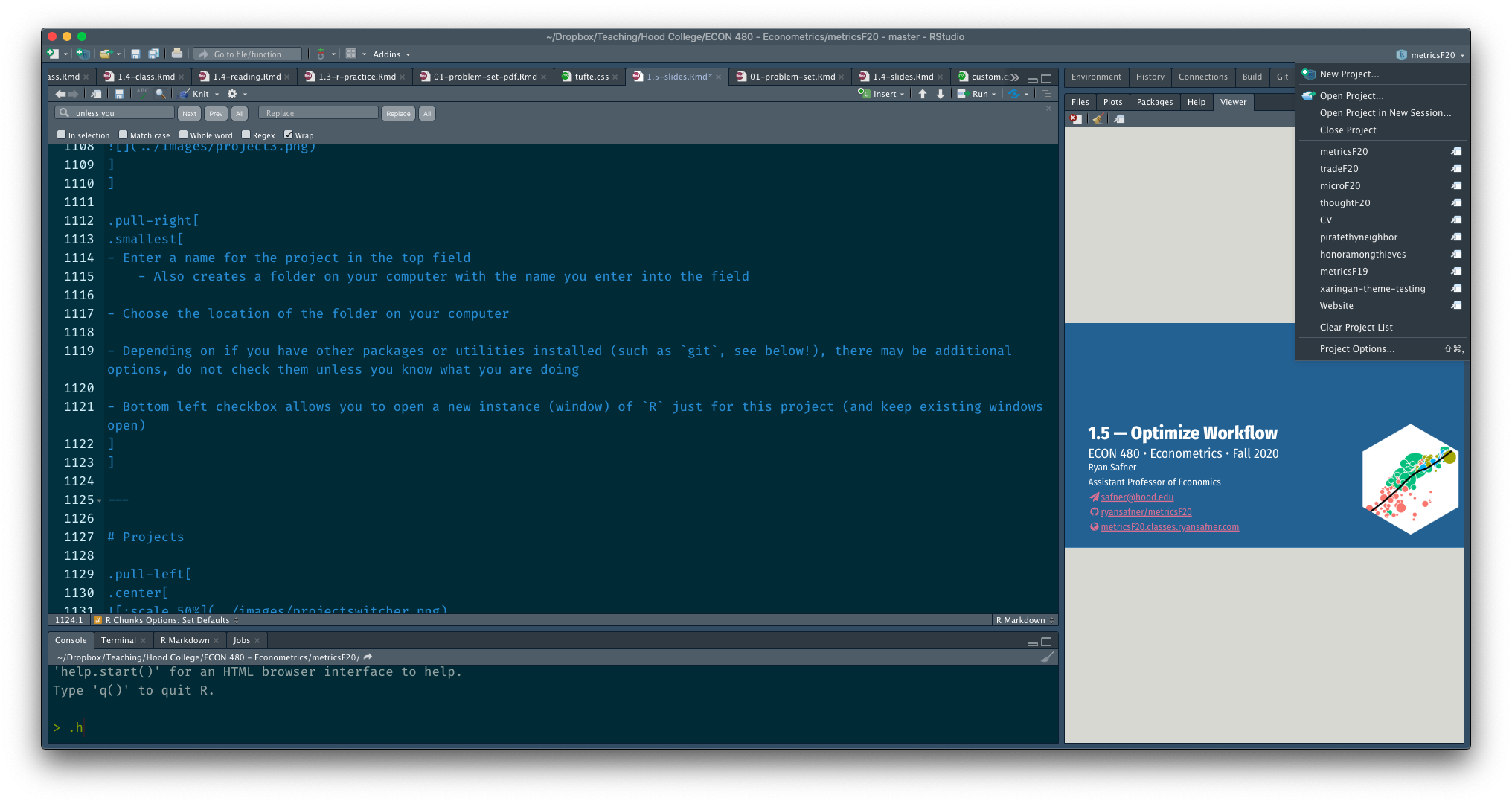
- At top right corner of RStudio
- Click the button to the right of the name to open in a new window!
Loading Others' Projects
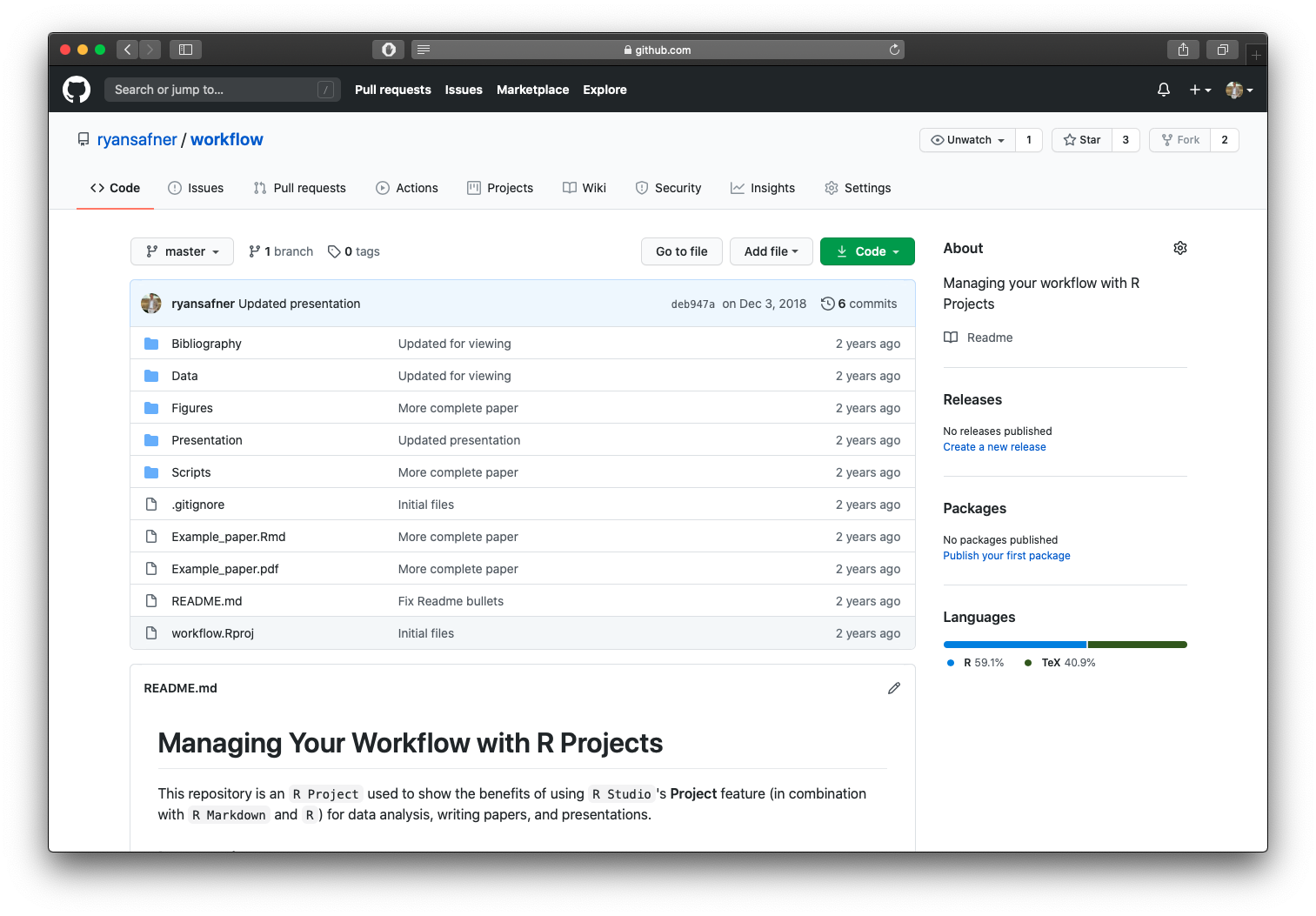
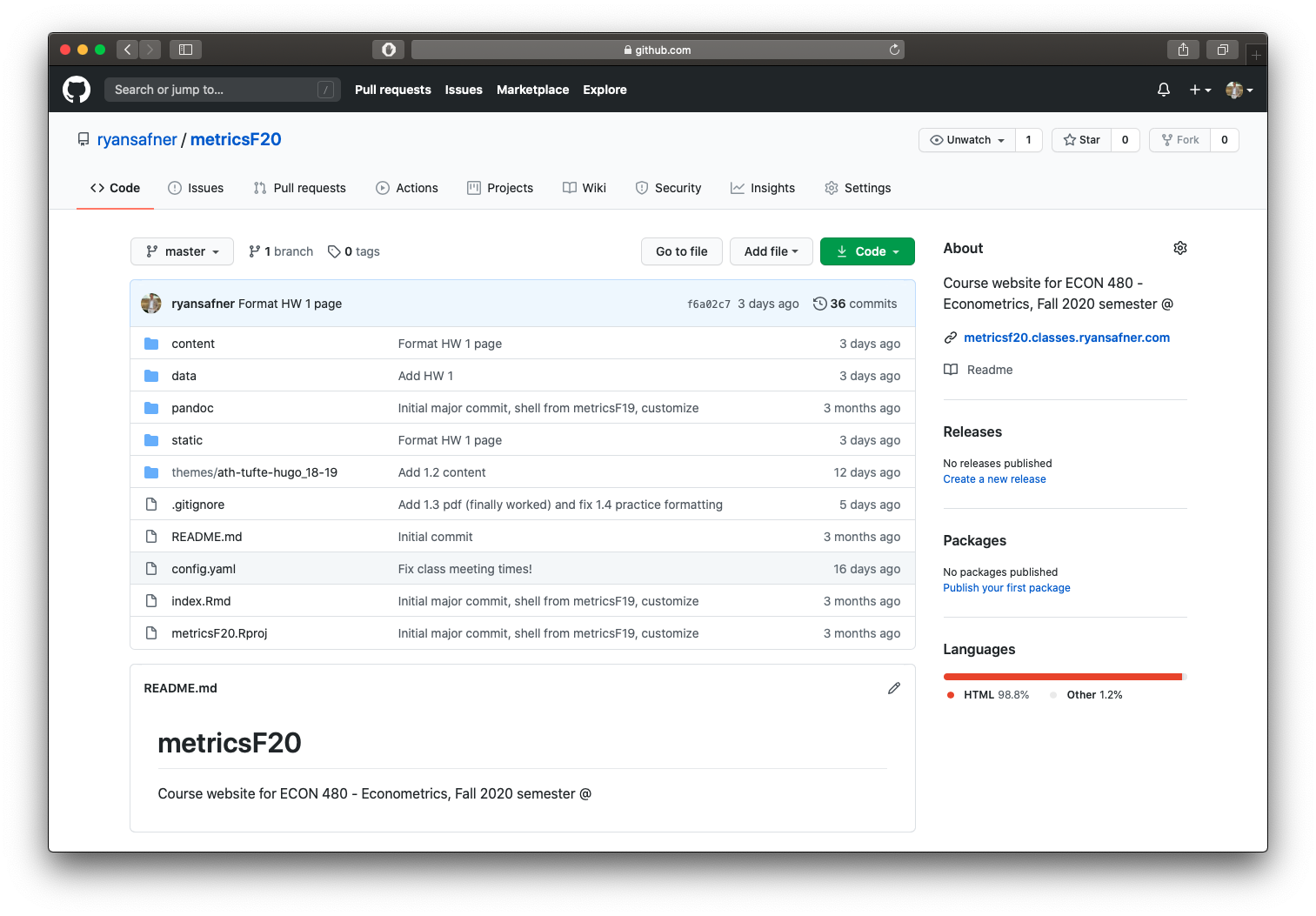
This project is on GitHub, click the green button, download to your computer, open .Rproj file in R Studio
A Good File Structure
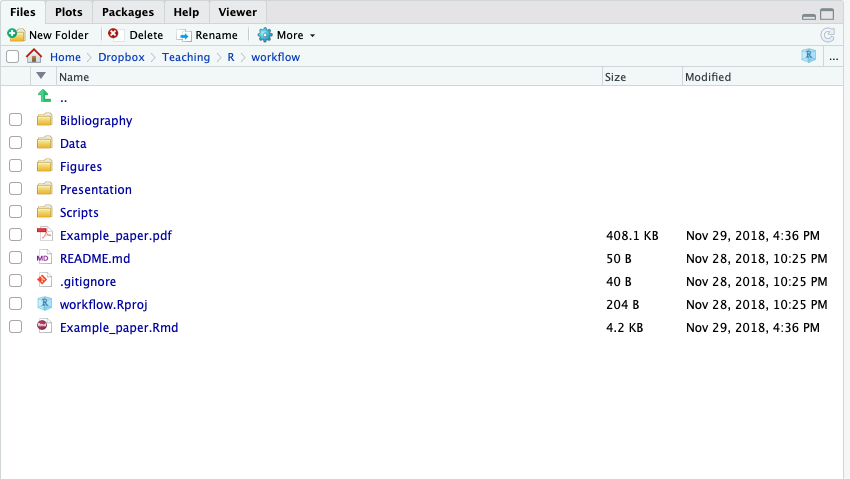
- Look through this on your own
- Read the
READMEof this repository on GitHub for instructions (automatically shows on the main page) - Look at the
Example_paper.Rmd- Uses data from Data folder
- Uses
.Rscripts from Scripts folder - Uses figures from Figures folder
- Uses
bibexample.bibfrom Bibliography folder
Version Control
Do You Want to Be Able To
Keep your files backed up
Track changes
Collaborate on the same files with others
Edit files on one computer and then open and continue working on another?

The Training-Wheels Version
Register an account for free
Set up a location on your computer for the
Dropbox/folderAnything you put in this folder will sync to the cloud
- As soon as you change files, they automatically update and sync!
- Can download any of these flies from the website on any device
- Set this up on multiple computers so when you change a file on one, it updates on all the others!
The Training-Wheels Version
My Dropbox - my life goes here
The Training-Wheels Version
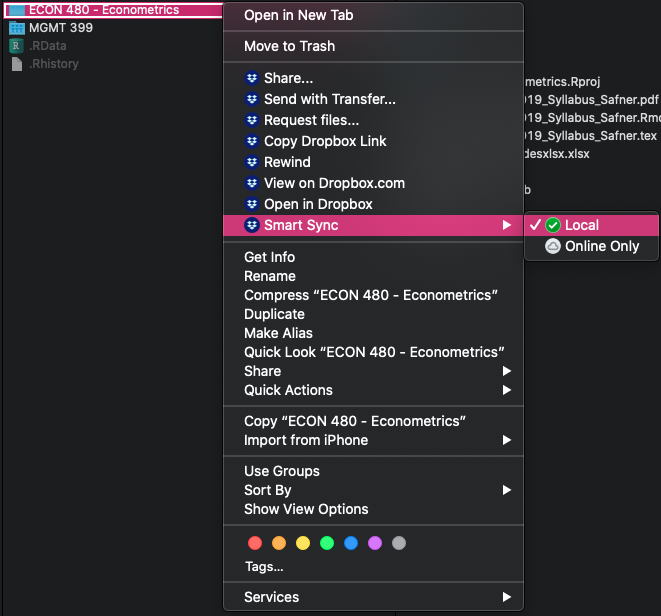
Smart Sync - keep some files online only for space
The Expert Version


Gitis an "open source distributed version control system" widely used in the software development industryTrack changes on steroids (if MS Word’s Track Changes and Dropbox had a baby)
- Organize folders/files to track (a
"repository") - Take a snapshot of all of your files (a "
commit") with "comments" pushthese to the cloudpullchanges to (other) computers as needed
- Organize folders/files to track (a
GitHubis a popular (not the only!) cloud destination for these repositories
The Expert Version
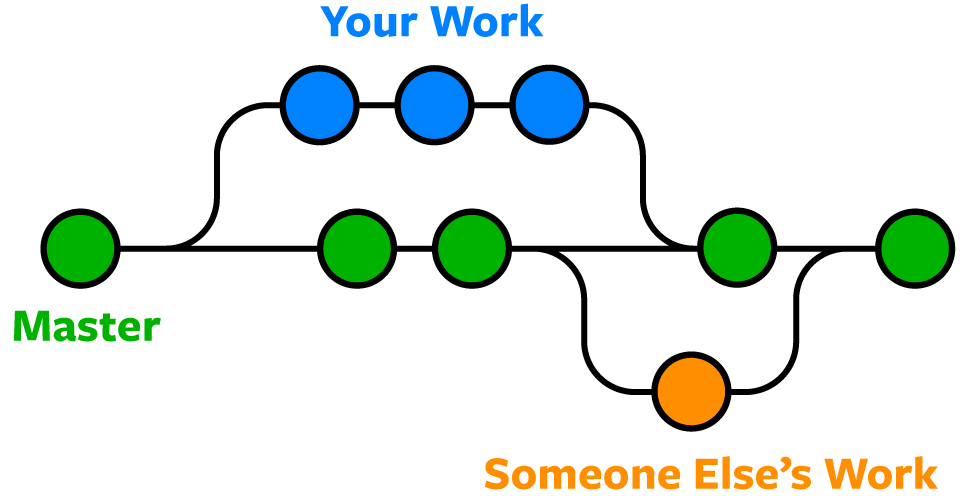
- Shows history (
versions) of files with comments- Can
forkorbranchrepository into multiple versions at once - Good for "testing" things out without destroying old versions!
revertback to original versions as needed
- Can
The Expert Version
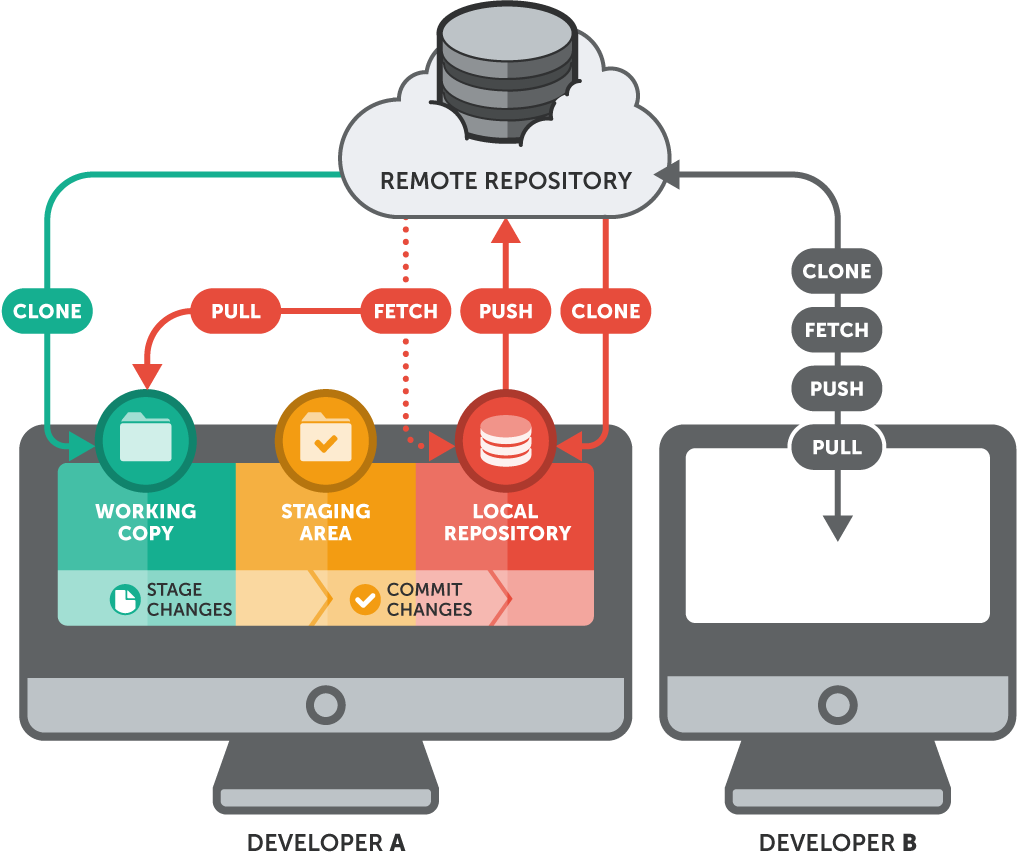
The Expert Version
Requires some advanced set up, see this excellent guide
R Studio integrates git and github commands nicely
My Workflow (that I suggest to you)
- Create a new repository on Github.*
- Start a New R Project in R Studio (link it to the github repository* - see guide)
- Create a logical file system (see example), such as:project # folder on my computer (the new working directory)||- Data/ # folder for data files|- Scripts/ # folder .R code|- Bibliography/ # folder for .bib files|- Figures/ # folder to plots and figures to|- paper.Rmd # write document here
- Write document in
paper.Rmd, loading/saving files from/to various folders in project- e.g. load data like
df<-read_csv("Data/my_data"); save plots likeggsave("Figures/p.png")
- e.g. load data like
- Knit document to
pdforhtml. - Occasionally,
stageandcommitchanges with a description,pushto GitHub.*
* Optional and a bit advanced, remember this is my workflow.
Resources
- R Studio's R Markdown Cheatsheet for a quick overview of R markdown
- R Studio's Overview of R Markdown for some tutorials
- R Studio's R Markdown Reference Guide for more specific options and issues
- Kieran Healey's The Plain Person's Guide to Plain Text Social Science on managing workflow with plain text files, R, and Git
- Yihui Xie's (and coauthors) R Markdown: the Definitive Guide on R Markdown syntax and customization options
- Hadley Wickham's (and Garrett Grolemund) R for Data Science on how to use R and R Markdown for data science work
- Jenny Bryan's Happy Git with R on how to use git and GitHub with R as a version control system