2.6 — Statistical Inference
ECON 480 • Econometrics • Fall 2021
Ryan Safner
Assistant Professor of Economics
safner@hood.edu
ryansafner/metricsF21
metricsF21.classes.ryansafner.com
Why Uncertainty Matters
Recall: The Two Big Problems with Data
- We use econometrics to identify causal relationships and make inferences about them
Problem for identification: endogeneity
- X is exogenous if cor(x,u)=0
- X is endogenous if cor(x,u)≠0
Problem for inference: randomness
- Data is random due to natural sampling variation
- Taking one sample of a population will yield slightly different information than another sample of the same population


Distributions of the OLS Estimators
OLS estimators (^β0 and ^β1) are computed from a finite (specific) sample of data
Our OLS model contains 2 sources of randomness:
Modeled randomness: u includes all factors affecting Y other than X
- different samples will have different values of those other factors (ui)
Sampling randomness: different samples will generate different OLS estimators
- Thus, ^β0,^β1 are also random variables, with their own sampling distribution
The Two Problems: Where We're Heading...Ultimately
Sample statistical inference→ Population causal indentification→ Unobserved Parameters
We want to identify causal relationships between population variables
- Logically first thing to consider
- Endogeneity problem
We'll use sample statistics to infer something about population parameters
- In practice, we'll only ever have a finite sample distribution of data
- We don't know the population distribution of data
- Randomness problem
Why Sample vs. Population Matters
Population
Why Sample vs. Population Matters

Population
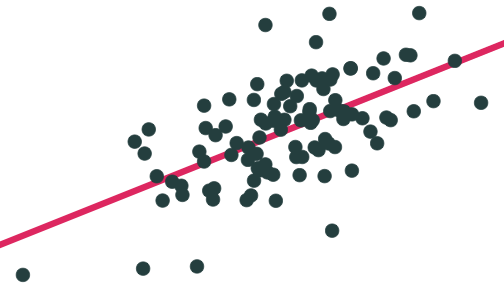
Population relationship
Yi=3.24+0.44Xi+ui
Yi=β0+β1Xi+ui
Why Sample vs. Population Matters
Sample 1: 30 random individuals
Why Sample vs. Population Matters

Sample 1: 30 random individuals
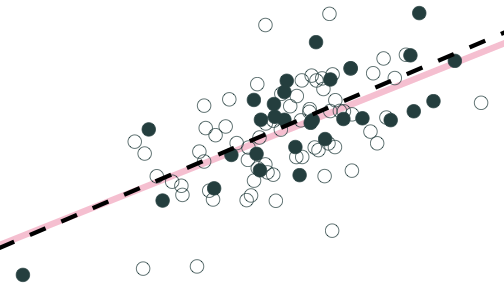
Population relationship
Yi=3.24+0.44Xi+ui
Sample relationship
ˆYi=3.19+0.47Xi
Why Sample vs. Population Matters
Sample 2: 30 random individuals
Population relationship
Yi=3.24+0.44Xi+ui
Sample relationship
ˆYi=4.26+0.25Xi
Why Sample vs. Population Matters
Sample 3: 30 random individuals
Population relationship
Yi=3.24+0.44Xi+ui
Sample relationship
ˆYi=2.91+0.46Xi
Why Sample vs. Population Matters
Let's repeat this process 10,000 times!
This exercise is called a (Monte Carlo) simulation
- I'll show you how to do this next class with the
inferpackage
- I'll show you how to do this next class with the
Why Sample vs. Population Matters
Let's repeat this process 10,000 times!
This exercise is called a (Monte Carlo) simulation
- I'll show you how to do this next class with the
inferpackage
- I'll show you how to do this next class with the
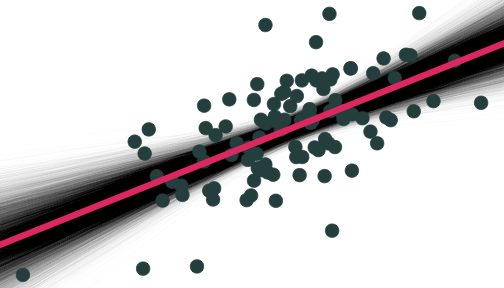
Why Sample vs. Population Matters
On average estimated regression lines from our hypothetical samples provide an unbiased estimate of the true population regression line E[^β1]=β1
However, any individual line (any one sample) can miss the mark
This leads to uncertainty about our estimated regression line
- Remember, we only have one sample in reality!
- This is why we care about the standard error of our line: se(^β1)!
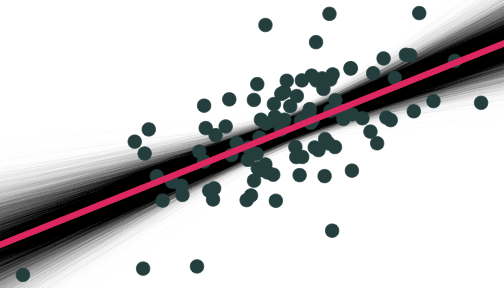
Confidence Intervals
Statistical Inference
Sample statistical inference→ Population causal indentification→ Unobserved Parameters
Statistical Inference
Sample statistical inference→ Population causal indentification→ Unobserved Parameters
- We want to start inferring what the true population regression model is, using our estimated regression model from our sample
Statistical Inference
Sample statistical inference→ Population causal indentification→ Unobserved Parameters
- We want to start inferring what the true population regression model is, using our estimated regression model from our sample
^Yi=^β0+^β1X🤞 hopefully 🤞→Yi=β0+β1X+ui
- We can’t yet make causal inferences about whether/how X causes Y
- coming after the midterm!
Estimation and Statistical Inference
Our problem with uncertainty is we don’t know whether our sample estimate is close or far from the unknown population parameter
But we can use our errors to learn how well our model statistics likely estimate the true parameters
Use ^β1 and its standard error, se(^β1) for statistical inference about true β1
We have two options...

Estimation and Statistical Inference

Point estimate
Use our ^β1 & se(^β1) to determine if statistically significant evidence to reject a hypothesized β1
Reporting a single value (^β1) is often not going to be the true population parameter (β1)

Confidence interval
- Use our ^β1 & se(^β1) to create a range of values that gives us a good chance of capturing the true β1
Accuracy vs. Precision

- More typical in econometrics to do hypothesis testing (next class)
Generating Confidence Intervals
We can generate our confidence interval by generating a “bootstrap” sampling distribution
This takes our sample data, and resamples it by selecting random observations with replacement
This allows us to approximate the sampling distribution of ^β1 by simulation!

Confidence Intervals Using the infer Package
Confidence Intervals Using the infer Package

- The
inferpackage allows you to do statistical inference in atidyway, following the philosophy of thetidyverse
# install first!install.packages("infer")# loadlibrary(infer)Confidence Intervals with the infer Package I

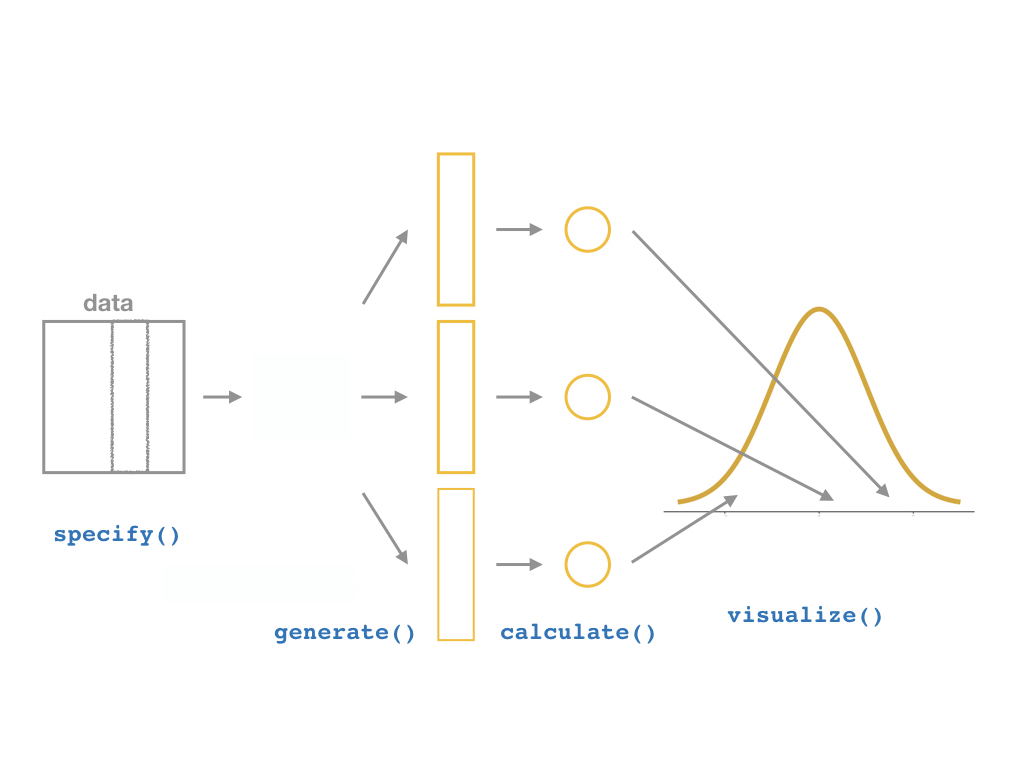
inferallows you to run through these steps manually to understand the process:
specify()a modelgenerate()a bootstrap distributioncalculate()the confidence intervalvisualize()with a histogram (optional)
Confidence Intervals with the infer Package II
Confidence Intervals with the infer Package II
Confidence Intervals with the infer Package II
Confidence Intervals with the infer Package II
Confidence Intervals with the infer Package II
Bootstrapping
Our Sample
| ABCDEFGHIJ0123456789 |
term <chr> | estimate <dbl> | std.error <dbl> | |
|---|---|---|---|
| (Intercept) | 698.932952 | 9.4674914 | |
| str | -2.279808 | 0.4798256 |
Bootstrapping
Our Sample
| ABCDEFGHIJ0123456789 |
term <chr> | estimate <dbl> | std.error <dbl> | |
|---|---|---|---|
| (Intercept) | 698.932952 | 9.4674914 | |
| str | -2.279808 | 0.4798256 |
Another “Sample”
| ABCDEFGHIJ0123456789 |
term <chr> | estimate <dbl> | std.error <dbl> | |
|---|---|---|---|
| (Intercept) | 708.270835 | 9.5041448 | |
| str | -2.797334 | 0.4802065 |
👆 Bootstrapped from Our Sample
Bootstrapping
Our Sample
| ABCDEFGHIJ0123456789 |
term <chr> | estimate <dbl> | std.error <dbl> | |
|---|---|---|---|
| (Intercept) | 698.932952 | 9.4674914 | |
| str | -2.279808 | 0.4798256 |
Another “Sample”
| ABCDEFGHIJ0123456789 |
term <chr> | estimate <dbl> | std.error <dbl> | |
|---|---|---|---|
| (Intercept) | 708.270835 | 9.5041448 | |
| str | -2.797334 | 0.4802065 |
👆 Bootstrapped from Our Sample
- Now we want to do this 1,000 times to simulate the unknown sampling distribution of ^β1
The infer Pipeline: Specify

The infer Pipeline: Specify
Specify
data %>%
specify(y ~ x)
- Take our data and pipe it into the
specify()function, which is essentially alm()function for regression (for our purposes)
CASchool %>% specify(testscr ~ str)| ABCDEFGHIJ0123456789 |
testscr <dbl> | str <dbl> | ||
|---|---|---|---|
| 690.80 | 17.88991 | ||
| 661.20 | 21.52466 | ||
| 643.60 | 18.69723 | ||
| 647.70 | 17.35714 | ||
| 640.85 | 18.67133 |
The infer Pipeline: Generate

The infer Pipeline: Generate
Specify
Generate
%>% generate(reps = n,
type = "bootstrap")
Now the magic starts, as we run a number of simulated samples
Set the number of
repsand settypeto"bootstrap"
CASchool %>% specify(testscr ~ str) %>% generate(reps = 1000, type = "bootstrap")The infer Pipeline: Generate
Specify
Generate
%>% generate(reps = n,
type = "bootstrap")
replicate: the “sample” number (1-1000)creates
xandyvalues (data points)
The infer Pipeline: Calculate
Specify
Generate
Calculate
%>% calculate(stat = "slope")
CASchool %>% specify(testscr ~ str) %>% generate(reps = 1000, type = "bootstrap") %>% calculate(stat = "slope")For each of the 1,000 replicates, calculate
slopeinlm(testscr ~ str)Calls it the
stat
The infer Pipeline: Calculate
Specify
Generate
Calculate
%>% calculate(stat = "slope")
boot <- CASchool %>% #<< # save this specify(testscr ~ str) %>% generate(reps = 1000, type = "bootstrap") %>% calculate(stat = "slope")bootis (our simulated) sampling distribution of ^β1!We can now use this to estimate the confidence interval from our ^β1=−2.28
And visualize it
Confidence Interval
- A 95% confidence interval is the middle 95% of the sampling distribution
| ABCDEFGHIJ0123456789 |
lower <dbl> | upper <dbl> | |
|---|---|---|
| -3.340545 | -1.238815 |
sampling_dist<-ggplot(data = boot)+ aes(x = stat)+ geom_histogram(color="white", fill = "#e64173")+ labs(x = expression(hat(beta[1])))+ theme_pander(base_family = "Fira Sans Condensed", base_size=20)sampling_dist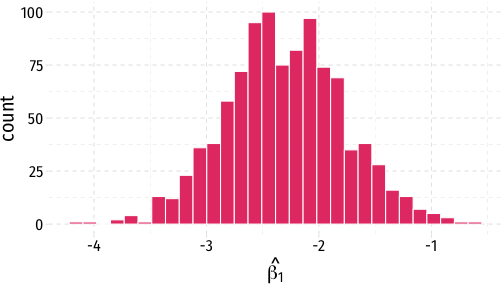
Confidence Interval
- A confidence interval is the middle 95% of the sampling distribution
ci<-boot %>% summarize(lower = quantile(stat, 0.025), upper = quantile(stat, 0.975))ci| ABCDEFGHIJ0123456789 |
lower <dbl> | upper <dbl> | |
|---|---|---|
| -3.340545 | -1.238815 |
sampling_dist+ geom_vline(data = ci, aes(xintercept = lower), size = 1, linetype="dashed")+ geom_vline(data = ci, aes(xintercept = upper), size = 1, linetype="dashed")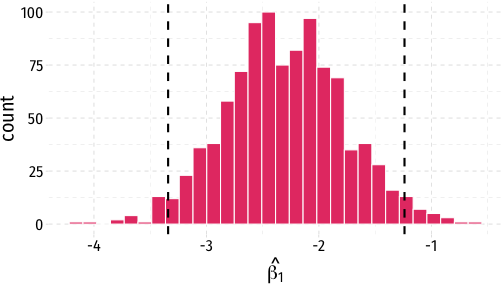
The infer Pipeline: Confidence Interval
Specify
Generate
Calculate
Get Confidence Interval
%>% get_confidence_interval()
CASchool %>% #<< # save this specify(testscr ~ str) %>% generate(reps = 1000, type = "bootstrap") %>% calculate(stat = "slope") %>% get_confidence_interval(level = 0.95, type = "se", point_estimate = -2.28)| ABCDEFGHIJ0123456789 |
lower_ci <dbl> | upper_ci <dbl> | ||
|---|---|---|---|
| -3.273376 | -1.286624 |
Broom Can Estimate a Confidence Interval
tidy_reg <- school_reg %>% tidy(conf.int = T)tidy_reg| ABCDEFGHIJ0123456789 |
term <chr> | estimate <dbl> | std.error <dbl> | statistic <dbl> | p.value <dbl> | conf.low <dbl> | conf.high <dbl> |
|---|---|---|---|---|---|---|
| (Intercept) | 698.932952 | 9.4674914 | 73.824514 | 6.569925e-242 | 680.32313 | 717.542779 |
| str | -2.279808 | 0.4798256 | -4.751327 | 2.783307e-06 | -3.22298 | -1.336637 |
Broom Can Estimate a Confidence Interval
tidy_reg <- school_reg %>% tidy(conf.int = T)tidy_reg| ABCDEFGHIJ0123456789 |
term <chr> | estimate <dbl> | std.error <dbl> | statistic <dbl> | p.value <dbl> | conf.low <dbl> | conf.high <dbl> |
|---|---|---|---|---|---|---|
| (Intercept) | 698.932952 | 9.4674914 | 73.824514 | 6.569925e-242 | 680.32313 | 717.542779 |
| str | -2.279808 | 0.4798256 | -4.751327 | 2.783307e-06 | -3.22298 | -1.336637 |
# save and extract confidence intervalour_CI <- tidy_reg %>% filter(term == "str") %>% select(conf.low, conf.high)our_CI| ABCDEFGHIJ0123456789 |
conf.low <dbl> | conf.high <dbl> | |||
|---|---|---|---|---|
| -3.22298 | -1.336637 |
The infer Pipeline: Confidence Interval
Specify
Generate
Calculate
Visualize
%>% visualize()
CASchool %>% #<< # save this specify(testscr ~ str) %>% generate(reps = 1000, type = "bootstrap") %>% calculate(stat = "slope") %>% visualize()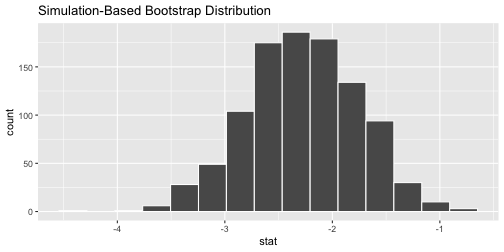
visualize()is just a wrapper forggplot()
The infer Pipeline: Confidence Interval
Specify
Generate
Calculate
Visualize
%>% visualize()
CASchool %>% #<< # save this specify(testscr ~ str) %>% generate(reps = 1000, type = "bootstrap") %>% calculate(stat = "slope") %>% visualize()+shade_ci(endpoints = our_CI)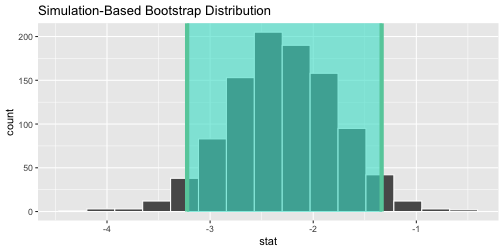
- If we have our confidence levels saved (
our_CI) we canshade_ci()ininfer'svisualize()function
Confidence Intervals
- In general, a confidence interval (CI) takes a point estimate and extrapolates it within some margin of error (MOE):
([ estimate - MOE ], [ estimate + MOE ])
- The main question is, how confident do we want to be that our interval contains the true parameter?
- Larger confidence level, larger margin of error (and thus larger interval)

Confidence Intervals
(1−α) is the confidence level of our confidence interval
- α is the “significance level” that we use in hypothesis testing
- α= probability that the true parameter is not contained within our interval
Typical levels: 90%, 95%, 99%
- 95% is especially common, α=0.05

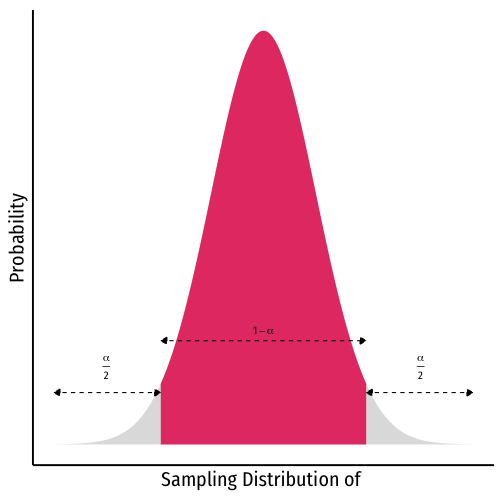
Confidence Levels
Depending on our confidence level, we are essentially looking for the middle (1−α)% of the sampling distribution
This puts α in the tails; α2 in each tail
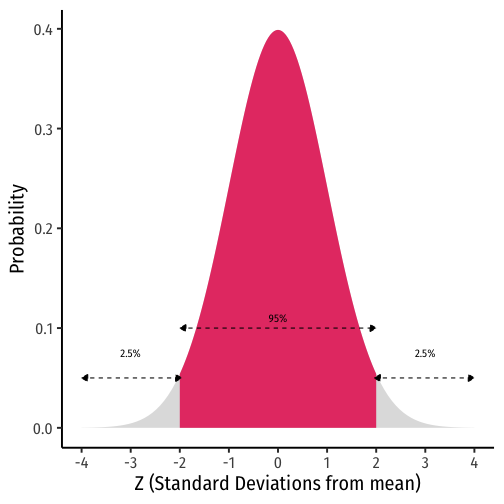
Confidence Levels and the Empirical Rule
Recall the 68-95-99.7% empirical rule for (standard) normal distributions!†
95% of data falls within 2 standard deviations of the mean
Thus, in 95% of samples, the true parameter is likely to fall within about 2 standard deviations of the sample estimate
† I’m playing fast and loose here, we can’t actually use the normal distribution, we use the Student’s t-distribution with n-k-1 degrees of freedom. But there’s no need to complicate things you don’t need to know about. Look at today’s class notes for more.
Interpreting Confidence Intervals
- So our confidence interval for our slope is (-3.22, -1.33), what does this mean again?
Interpreting Confidence Intervals
- So our confidence interval for our slope is (-3.22, -1.33), what does this mean again?
❌ 95% of the time, the true effect of class size on test score will be between -3.22 and -1.33
Interpreting Confidence Intervals
- So our confidence interval for our slope is (-3.22, -1.33), what does this mean again?
❌ 95% of the time, the true effect of class size on test score will be between -3.22 and -1.33
❌ We are 95% confident that a randomly selected school district will have an effect of class size on test score between -3.22 and -1.33
Interpreting Confidence Intervals
- So our confidence interval for our slope is (-3.22, -1.33), what does this mean again?
❌ 95% of the time, the true effect of class size on test score will be between -3.22 and -1.33
❌ We are 95% confident that a randomly selected school district will have an effect of class size on test score between -3.22 and -1.33
❌ The effect of class size on test score is -2.28 95% of the time.
Interpreting Confidence Intervals
- So our confidence interval for our slope is (-3.22, -1.33), what does this mean again?
❌ 95% of the time, the true effect of class size on test score will be between -3.22 and -1.33
❌ We are 95% confident that a randomly selected school district will have an effect of class size on test score between -3.22 and -1.33
❌ The effect of class size on test score is -2.28 95% of the time.
✅ We are 95% confident that in similarly constructed samples, the true effect is between -3.22 and -1.33
Estimating in R (Via Regression, rather than infer)
- Base R doesn't show confidence intervals in the
lm summary()output, need theconfintcommand
confint(school_reg)## 2.5 % 97.5 %## (Intercept) 680.32313 717.542779## str -3.22298 -1.336637broomcan include confidence intervals
school_reg %>% tidy(conf.int = TRUE)| ABCDEFGHIJ0123456789 |
term <chr> | estimate <dbl> | |
|---|---|---|
| (Intercept) | 698.932952 | |
| str | -2.279808 |